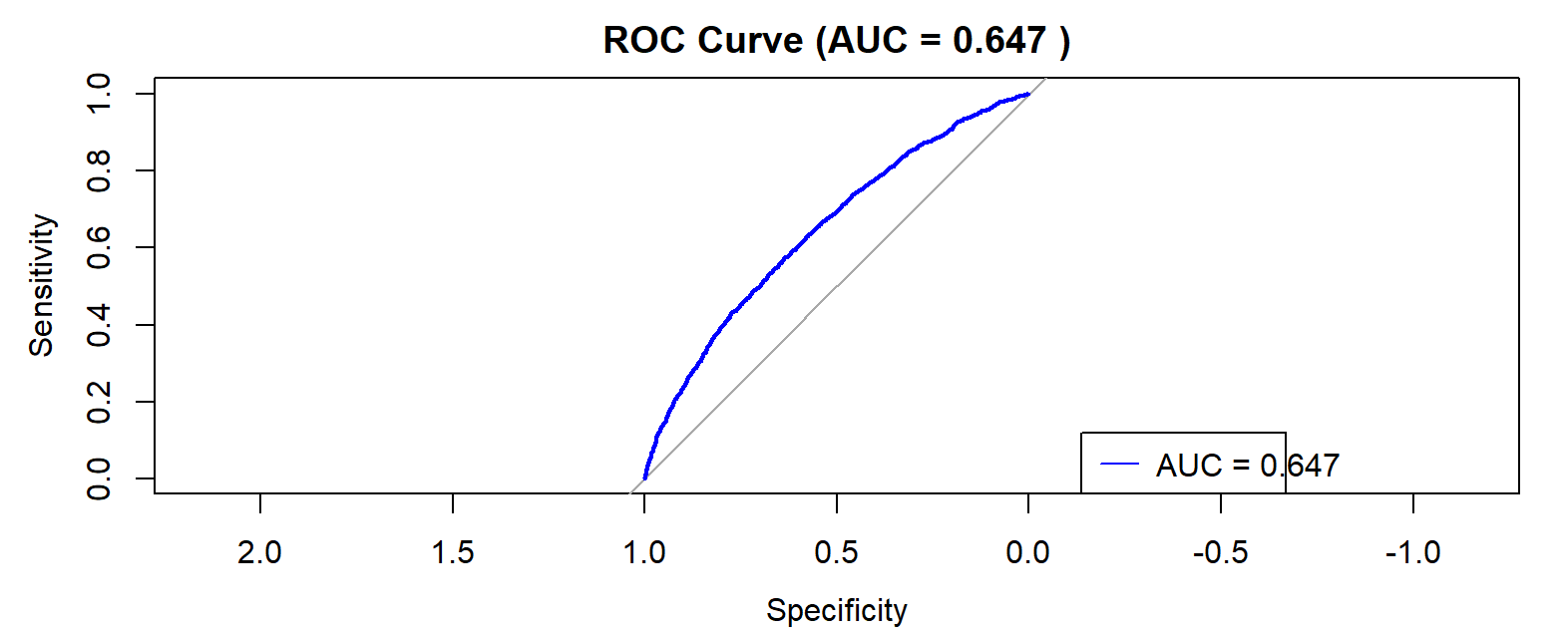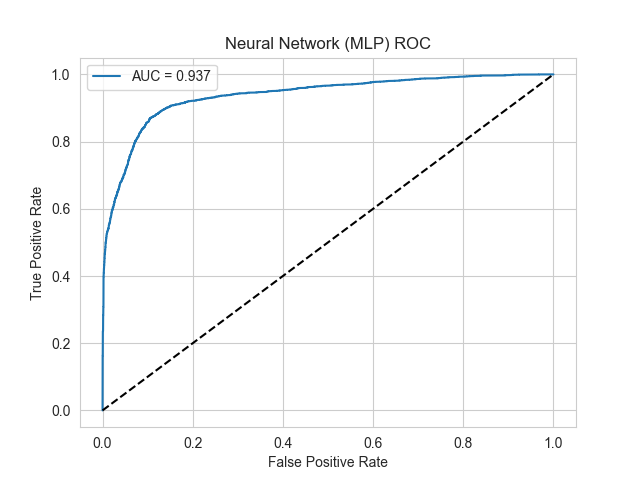
Glioblastoma is the most malignant primary brain tumor with high heterogeneity, making it challenging to achieve accurate diagnosis and evaluate treatment efficacy. With the fast development of single-cell RNA sequencing technology, malignant cells can be identified at the single-cell level to evaluate tumor purity. This study developed a computational workflow that integrated single-cell sequencing data and machine learning methods. Two classification models, XGBoost and a multilayer perceptron, were developed based on 30 selected genes with most differential expression identified by an independent samples t-tests from whole-genome expression data. Subsequently, the performance of two models was evaluated using multiple evaluation metrics. Experimental results showed that the two machine learning models had excellent performance in distinguishing malignant cells in glioblastoma. For distinguishing malignant cells, the AUC, accuracy, sensitivity and specificity of the XGBoost model were 0.941, 0.894, 0.883 and 0.901, respectively; while those of the MLP model were 0.937, 0.883, 0.865 and 0.896, respectively. In addition, the results of the probability distribution experiment showed that the XGBoost model had a more concentrated distribution, while the MLP model had a relatively broader distribution. These results were consistent with the effectiveness of the two machine learning approaches in malignant cell identification. This study validated the effectiveness of using machine learning methods based on single-cell RNA-seq data in identifying malignant cells in glioblastoma. This machine learning workflow could provide a reliable computational tool for subsequent malignant cell identification and tumor purity assessment.